Molecular Dynamics Study of Calmodulin in Potassium Channels
Internship
Type of Project: Theory
Location: Donostia
Supervisors:
Aitor Bergara
Aritz Leonardo
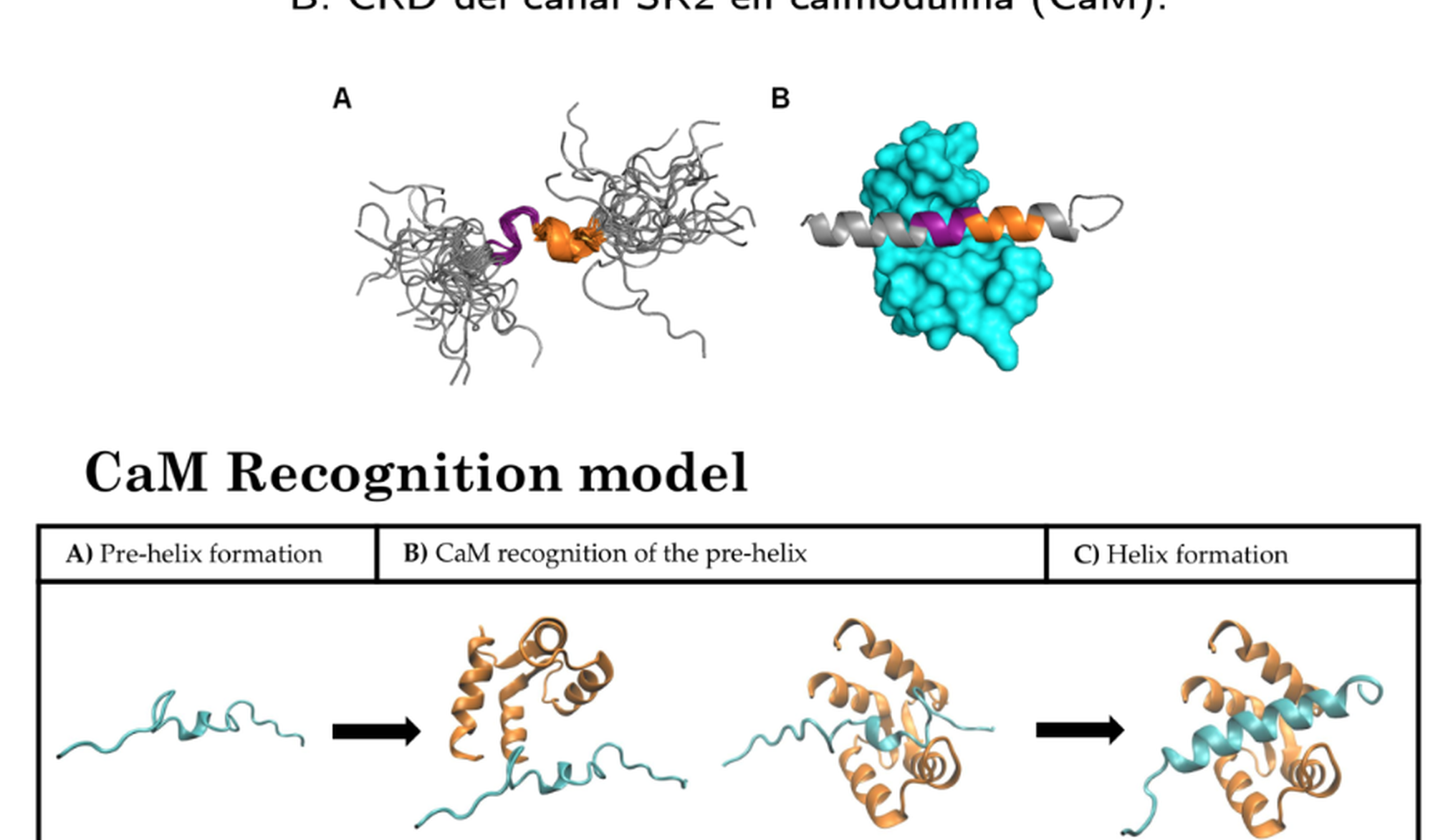
The complete resolution of the sequence of the human genome in 2016 has been a huge milestone that continuously reveals causal relations among pathologies and gene signaling. Moreover, the latest advances of experimental techniques, such as nuclear magnetic resonance, provide a direct access to 3D maps with atomic resolution of the proteins that form cell membranes. This new accessible structural information has become a revolution in biological sciences for the design of new drugs that improve the well-being of humans. Remarkably, 60% of the commercial drugs act precisely on the proteins located at the cell membranes. With this scenario in mind, physical models that mimic atomic interactions within the proteins and their posterior time evolution through molecular dynamics provide a very powerful tool for the prediction and comprehension of membrane phenomena. Theoretical simulations of proteins serve as guidance for the design of new drugs and help understanding the enormous amount of experimental information available. In this project we shall consider the Kv7.2 channel of neuron membranes – a potassium voltage-gated channel located in human neurons – whose functioning relies in a potential difference induced by calmodulin (CaM). By means of all-atom simulations, molecular dynamics and coarse grain models, we would like to validate two claims regarding the behavior of the secondary structure of the IQ motiv of the channel:
1) Both wild type and mutant can form stable helices without the ribosome;
2) Wild type can form an alpha helix at the ribosome whereas the mutant cannot.
